Poly (A) Tails: does this protein make my tail look fat?
Lots of ongoing questions, few answers on this little talked about part of the mRNA molecule
Well, I wish I could take credit for that catchy headline but it is actually from a paper that discussed the determinant and implication of Poly (A) tail size (see here)
We don't talk much about the poly(A) tails in the biosynthetic modRNA but we should. It is considered a critical quality attribute, important to the production of the spike protein. But there are many issues with the poly (A) tail that continue to cause issues for the manufacturers.
transcribing the poly(A) tail from the DNA template results in modRNA with various tail lengths
no one knows how to measure it
The modRNA
A review of the modRNA structure. The poly (A) tails are a is a long chain of adenine nucleotides that is added to a messenger RNA (mRNA) molecule during RNA processing to increase the stability of the molecule. They are normally about 100-250 residues long. Inside a cell, processing of the 3' end adds a poly-A tail to the RNA molecule with an enzyme called poly-A polymerase in a process called polyadenylation. But since we are doing in vitro transcription, certain other issues arise as we will discuss below.
The entire modRNA produced by in vitro transcription must be tested for and meet certain quality attributes for
identity
purity
stability
immunogenicity
homogenicity
Here is the modRNA and its relevant parts.
DETAILS OF THE POLY A TAILS IN THE modRNA JABS
And here is another visualization which includes detail on the Poly A tails (which are segmented as discussed below). For the Pfizer/BioNTech vaccine it is:
30 adenosines
A 10 nucleotide linker sequence
followed by another 70 adenosine residues.
The total poly (A) tail is 110nt in length
Image below is from this paper
IN VITRO TRANSCRIPTION OF THE POLY A TAILS
Now with in vitro transcription there are 3 ways of adding the poly A tails.
Poly (A) tails can be added to the modRNA by
added via PCR (not shown above)
added via enzymatic polyadenylation after IVT (see above = Process 1) but might result in heterogenous tails
included in the template plasmid (Process 2)
Above is a great graphic from this excellent paper which shows you how Pfizer manufactured Process 1 and Process 2 modRNA. For the purposes of this substack, it is the lower portion which shows how the poly A tail is encoded in the plasmid DNA , whereas Process 1 adds the poly A tail to the mRNA enzymatically.
The Poly (A) tail was included in the plasmid for Moderna which used a plasmid and a Process 2 manufacturing from the very beginning. Both add the 5’cap to the mRNA though.
So you can imagine there are issues with transcribing the polyA tails directly from the plasmid. These comments are from the EMA leak/hack
Unfortunately the EMA document does not include this “investigation” by Pfizer/BioNTech.
What do Poly (A) tails do?
I am no molecular biologist, but everything I read is that the poly (A) tails are essential, do a lot of stuff, do stuff we don’t yet know, and regulate gene expression. Also poly (A) tails are found in almost all mRNA eukaryote cells so they must be important.
They are needed for:
stability
protects RNA from degradation
used in translation initiation
influences translation efficacy
So pretty important.
Lets talk segmented poly A tails
What are the factors affecting Poly A tails and translation/stability?
Mostly it is about short tails vs long tails and optimal length for the modRNA in the vaccines. For the modRNA jabs they are 110nt long. There are a bunch of other issues that is too much for my puny brain but can be reviewed in that catchy title paper here.
BUT WHAT ABOUT THESE SEGMENTED POLY (A) TAILS IN THE JABS?
segmented poly A tails as found in the modRNA of the jabs
this is a biosynthetic modification as segmented poly A tails are not found in nature
results in higher protein production (have to have LOTS of spike protein)
for process 2 jabs of Pfizer and for Moderna, plasmids which encode the poly (A) tails can recombine in E coli and then you’d have shortened tails. Segmented poly A tails reduces this recombination without affecting mRNA half life (often increasing it) or in protein production. MORE INFO HERE
actually I found a GOOGLE PATENT that explains these segmented poly (A) tails well and why this was ANOTHER chosen synthetic modification of mRNA
GOOGLE PATENT ON SEGMENTED POLY (A) TAILS
This was filed by ETHRIS GMBH a start up in Germany in 2020
Of note, the EMA leak/hack states this:
Translational efficacy in dendritic cells??? Hmmmm…..how long did it take their scientists to figure this out?
How are they degraded?
NORMAL mRNA
Like a good pharmacologist I always ask how does the body get rid of the mRNA? Well it starts by breaking down the Poly (A) tails, called depolyadenylation. This will become important later. Pic is of normal mRNA
So how I understand it, from this paper the Poly (A) tails are removed in small chunks, then the 5’ cap, then the rest of the mRNA degrades. So degradation of the mRNA starts with the poly (A) tail.
Segmented Poly (A)Tailed modRNA
Well, this is hard to find, of course since it is all proprietary and it is not even mentioned in the patent above. I would assume the degradation of the mRNA would be much slower given these segmented poly A tails. I think this may be a possible explanation or partial explanation of why the synthetic mRNA lasts so long. Course, it could also be protected in exosomes.
Well here we go. In this paper by Pilkington we find this:
Well, you don’t say.
But wait! There’s more! It appears that the modRNA from the jabs can be REpolyadenylated. But this ONLY happens IN VIVO and not in vitro. Well that is another way the modRNA can hang around for a while and not be degraded.
This comes from this rather infamous paper, published as a preprint in Dec 2022
SARS-CoV-2 mRNA vaccine is re-adenylated in vivo, enhancing antigen production and immune response
Authors: Paweł S Krawczyk1, et al
They showed, that using the Moderna jab:
Notably, mRNA-1273 molecules are re-adenylated after mΨCmΨAG removal. Detailed analysis of immune cells involved in antigen production revealed that in macrophages, after mΨCmΨAG removal, vaccine mRNA is very efficiently re-adenylated, and poly(A) tails can reach up to 200A. In contrast, in dendritic cells, vaccine mRNA undergoes slow deadenylation-dependent decay. We further demonstrate that enhancement of mRNA stability in macrophages is mediated by TENT5 poly(A) polymerases, whose expression is induced by the vaccine itself.
Wow. Many of us on twitter thought this was a pretty important finding. Note the slow deadylation in denditric cells as stated in the EMA leak/hack AND REpolyadenylation elsewhere.
Most interesting, this paper was never published, despite the intense interest.
Curiouser and Curiouser.
HOW ARE POLY (A) TAILS PROFILED AND MEASURED?
OK this is where it gets very very interesting. So we know poly A tails are important, they are considered a critical quality attribute, so you need to measure them and ensure you have the complete poly A tail attached to the mRNA. Otherwise, you might not get translation, or there maybe other issues.
At a recent USP symposium I attended, there were 3 presentations of how the measure the poly A tails for
tail length
tail distribution
which are the critical quality attributes.
So the potential analytical methods for testing for Poly A includes:
gel elecrophoresis
sequencing
capillary elecrophoris
liquid chromatography including
size exclusion chromatography
anion-exchange chromatography
reverse phase chromatography
RP-LC (reverse phase liquid chromatography)
IP-RP-LC (ion phase RP-LC)
LC/MS (liquid chromatography/mass spec)
an example of LC/MS results from the batch comparison of the Pfizer jab as found in the EMA leak/hack
The 20Y513C101 is a Process 2 batch, the others are Process 1. As you can see there is a decrease in homogeneity of the L70 segment of the Poly (A) tail with Process 2 as discussed above.
What are the compendial standards for measuring the poly (A) tails going to be?
So a bunch of analytical chemists and biopharm scientists from a wide variety of companies called BioPhorum (those supplying raw ingredients, specialized equipment, contractors, and the drug companies themselves), got together to discuss this and other standards. Their analysis can be found HERE and it is worthwhile downloading the document and reading the commentary.
Under purity, they suggest these analytical methods for the poly (A) tails
HOWEVER, the second draft USP Guidelines only suggests IPRP-LC as the compendial standard
Measuring Poly (A) tails and determining a compendial standard is still in flux, based on what I am reading and on the USP Open Forum I attended. There were 3 presentations on the different ways Poly (A) tail can be measured.
Pfizer/BioNTech Tries to Fool the EMA on measuring the Poly (A) tails
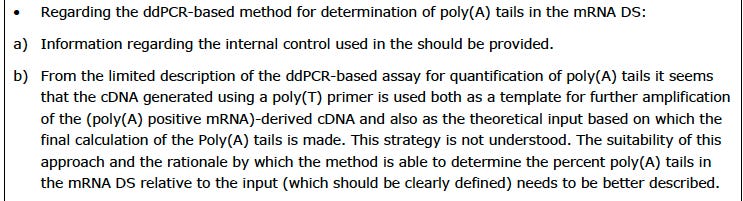
Pfizer/BioNTech didn’t use any of the analytical methods listed above. They used something called ddPCR. Lets see what the EMA leak/hack said about BioNTech using ddPCR for measuring the poly (A) tails.
This strategy IS NOT UNDERSTOOD. Yeah. There are 2 instances of this in the EMA document.
using excess CTP in the IVT of the modRNA which results in less dsRNA but a lot more dsDNA (see my substack entry on this topic)
using ddPCR for measuring poly (A) tails.
What kind of results did they get using ddPCR?
Did you catch the issue here?
BioNTech used ANOTHER PROCESS to measure poly (A) tails in process 1 jabs but changed to ddPCR in the Process 2 jabs. What was it? It was IPRP-LC, which is the method recommended for both tail length and distribution AS WELL as percent of mRNAs having poly(A) tails. They also appeared to use LC/MS to compare Process 1 with Process 2. But they dropped both methods for the commercial Process 2 lots. WHY?
The EMA caught on to this:
As of today they are still using ddPCR for the poly (A) tails and have tried to justify it with this analysis which references the infamous Patel paper. They call the Poly (A) tails the ACHILLES HEEL of modRNA quality.
But why did they change from Process 1 to Process 2? Because they got better values? Other reasons? Look what we found out about the excess CTP in the IVT.
Anyone have any ideas? I asked the USP coordinator for her thoughts on this and if ddPCR was going to be used as a compendial standard and it was a flat NO, notwithstanding what the BioPhorum scientists say (they want to sell those fancy equipment after all).
Curiouser and curiouser.
SUMMARY
Poly (A) tails are important in modRNA and are considered the Achilles heel of mRNA production
The poly (A) tails in the modRNA are segmented and biosynthetic since they don’t appear in nature. (please let me know if I am wrong)
Segmented poly (A) tails seem to: (any maybe this explains the longevity of the mRNA in human tissue?)
target dendritic cells
slow degradation
can be REpolyadenylated
Compendial standards for poly (A) tails are evolving but seem to consist of measuring homogeneity (tail length and distribution) as well as percent of mRNA with a poly (A) tail
Pfizer/BioNTech is using ddPCR technology for both these CQAs (I think!) or at least for the percent of mRNA with a tail, but it is not certain, even after 4 years, that it is measuring what it is supposed to
The scientists at the coal face of manufacturing these mRNA products admit they do not know what to measure, how to measure and what these jabs are doing, especially the LNPs.
I love this (from the BioPharm collaboration):
Yes, this mRNA production is still an immature field with fatal flaws. These scientists are stating that they don’t even understand WHAT is important in measuring the critical quality attributes, NOR HOW to measure them since the techniques available are probably not good enough even after 4 years of intense study and lots and lots of $$$. I think they were all caught up in the novelty and promise of using mRNA as a therapeutic.
Reality is settling in.
Thanks for reading! And pray the rosary




















As always, thank you! So many details and clear information. I began to analyze the poly(A) tail issue in my book and it quickly became clear how little we know about the entire RNA life-cycle. So many things going the opposite way then told in the narrative. It is sobering to see, as you describe, that many of the issues and gaps were known during "vaccine" "design" and rollout.
Thanks Maria. Your readers might already know that Pfizer Process 2 did not apply the purification techniques shown in the Figure in the paper by Anna Blakney and coworkers. Pfizer admitted in March 2021, in a correction, initially lying to the FDA and EMA in November 2020 by claiming Endotoxin is controlled, whereas it is only measured in every batch. The LAL test EU/ml results are deadly secrets.
https://geoffpain.substack.com/p/production-of-the-pfizer-biontech