Testing and Analyzing the modRNA Vaccines; mRNA drug product
How can we determine harm if we don't even know what or how to analyze these so called vaccines?
I think I am one of the few people in the western world who has read that entire 190 page leaked review of the mRNA manufacturing, at least 30 times. Plus some of the other data that was leaked.
One aspect not talked about a lot, and was a huge concern for the EMA regulators was validating the analytical methods they used to test the mRNA vaccines for purity and quality. How do you know they are pure and of high quality if your analytical techniques are not? And more fundamentally, are we measuring what we need to measure and with the right tools?
Changes in Analytical Methods During the Development of the vaccines
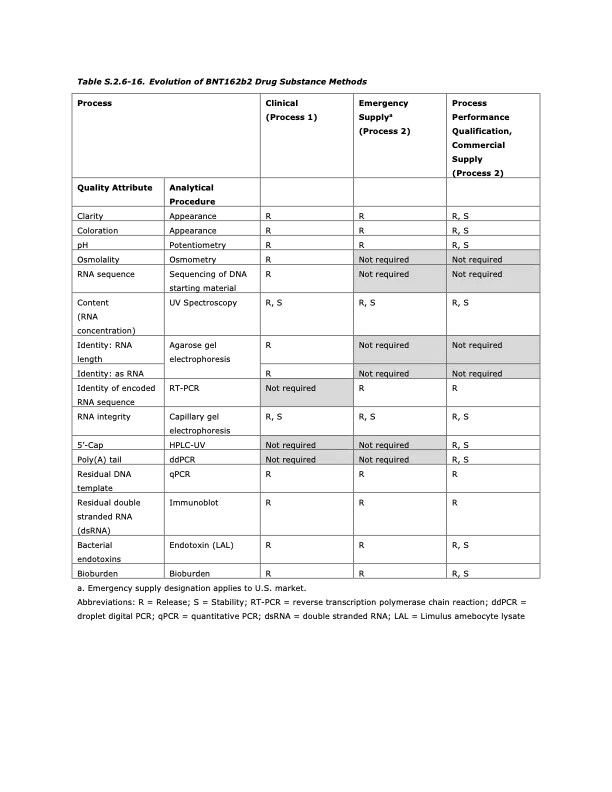
It is very instructive to look at how Process 1 lots were tested compared to process 2 lots, what was changed and what was kept. In addition, what parameters should be measured?? These are the critical quality attributes. Who decides what they are? Generally it is the manufacturers since they know how to make the product, but the regulators can ask for more testing and more quality attributes if there is a potential issue.
Process 1 vs Process 2 for modRNA drug substance
OK, can everyone see what changed between Process 1 and 2?
RNA identity of the mRNA
-in other words, was the right mRNA made from the DNA template? In process 1 they sequenced the DNA template for each batch which should be faithfully transcribed to the mRNA (and generally it is with only 1:1000 base pair errors or so). But for process 2 they are using RT-PCR. What are the implications? Do the primers used cover the entire mRNA strand?
Most importantly, since the DNA template is no longer sequenced, the SV40 promoter/enhancer as part of the the DNA template (which wasnt in Process 1 DNA template) is NOT MEASURED in the process 2 lots. So that’s how the regulators were blind to the deception. It is not unreasonable to switch to RT-PCR for the mRNA sequence verification if you are making lots and lots of product, so no suspicion was raised. But each batch of DNA template should be sequenced to ensure accuracy.
Poly (A) tail
In the process 1 lots they used HPLC and other methods, but switched to something called digital droplet PCR for the process 2 lots. This caused a lot of consternation to the EMA reviewers because this analytical method was never used for measuring the poly (A) tail. I have been digging into that for a long time with no clear answer, except that Pfizer/BioNTech insist on using this. So I think there is something going on with the 3’UTR and the tails, that I do not understand.
IF we look at the last column there is a total of 12 tests that must be done on the mRNA to determine purity and quality, or critical quality attributes at the time these products were released onto the market.
Fast forward 3.5 years.
USP compendial standards for the mRNA drug substance
I talk about the United States Pharmacopoeia here and their role in determining the actual analytical method to be used in testing AND the steps in performing this method. I think my colleague David Wiseman and myself are the few non industry, non academic people who have attended the training courses and forums that are offered by USP. And boy, we have learned a lot.
This is where you learn how the sausage is made, these are the engineers, chemists, and formulation specialists doing the actual work. They are honest about the challenges, and they are trying to make a reliable, quality product. These scientists are really siloed and likely have no idea on what the mRNA actually does or can do, or are driven by trying to clean it up because they think this technology will be useful for cancer.
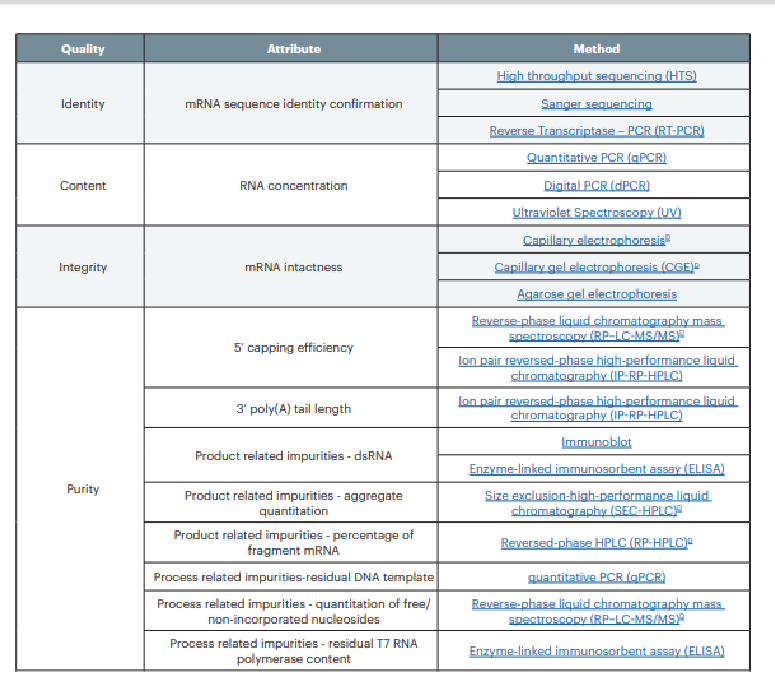
OK lets look at what the USP believes are the quality attributes that must be measured and what analytical tools to measure them.
Do you see they have increase from 12 critical quality attributes to…..
17 CQAs???
Proposed Critical Quality Attributes by USP
OK lets analyze this
There are several methods for many of these to accommodate differences in testing equipment
identity: yeah they want sequencing first, then maybe RT-PCR as the least sensitive method? So this is a definite improvement.
content: from JUST UV spectroscopy to ddPCR and qPCR which are much more sensitive methods. Addresses Kevin McKernan’s concerns re PCR for DNA but not for RNA. Both will be measured by qPCR at least for the mRNA drug substance before it gets shipped off to be put into the LNPs (but maybe not in the final product)
integrity (%intact mRNA): from CGP to AGE and capillary elecrophoresis but this is essentially unchanged
5’cap efficiency: from HPLC to very sensitive IP-RP-HPLC or RP-LC-MS/MS. Ha. Only an mRNA with a 5’ cap can be translated. So maybe they were undermeasuring how many mRNAs had the 5’cap? I think that is a possibility.
3’ UTR and poly A tail: uh oh NO ddPCR!!!! They want IP-RP-HPLC. What will Pfizer do? I actually asked the USP head of global biologics if they will accept ddPCR for measuring the poly(A) tails. SHE HAD A FLAT NO. Meanwhile Pfizer is writing articles on how good ddPCR is for the poly (A) tail. HMMMMM
dsRNA: there are no good tests for this. In fact, that is something some equipment manufacturers are working on because neither the immunoblot, or the ELISA methods are very good at determining in vivo effects. And dsRNA contamination is a very serious issue that more people needs to know.
Product-related impurities
DNA: USP does not want to change from qPCR. We asked them. Not exactly sure why. But they were fully aware of the controversy. I think as an NGO that provides these standards to the FDA, I dont think they want to rock the boat.
aggegate quantification: this is NEW!! what does this mean? RNA/DNA hybrids? Stable -folded RNA? other RNA impurities like loop-back RNA? So these are importan impurities that were not measured IN THE FIRST PLACE.
quantification of free, non-incorporated nucleosides. UH OH. You mean like free N-1-methypseudouridine? That on ITS OWN may be a driver for cancer? Why was this not considered before???
residual T7 polymerase: you mean having an bacterial polymerase in our cells which have been transfected IS NOT A GOOD IDEA? Who knew? Why was this not considered before?
Potency: cell based assay. This is a whole new substack because it is a very big problem. And BionTech admitted it and neither company really know how to make the potency determination, a reproducible test which reflects what is going on in the body.
So now we have new contaminants we need to measure for purity!
Not only that, but the quality of the starting materials need to be good, as we found out from the DNAse that breaks down the DNA template. Different vendors have difficult purity and efficacy parameters.
Here for example is the T7 polymerase
unicorporated nucleosides
Here is how you measure specific nucleoside bases in the mRNA. Of course this wasnt done with Process 1 or 2 lots, but it is going to be added to the analytical testing.
Base composition by LC provides specific details regarding the base composition of the sequences and how does it match to the theoretical sequence base composition. You can see errors in mRNA sequencing and excess nucleosides.
Oh but it gets worse. Here is a great analysis of what I have been writing about, indirectly shown with the lipid adducts. It is the stability of the mRNA and base modifications which making the mRNA, storing it, making in into the LNPs, and freezing it. METAL IONS in the LNPs. Another contamination. Figure out if the mRNA is even making spike protein (or other protein). These lipid adducts and stable folded mRNA IS A VERY BIG DEAL. How come no one else is talking about it?
aggregates? this is mostly RNA aggregates. We need DNA as well.
Well they talk about how it impacts therapeutic efficacy (TE). But what about adverse events? We know aldehydes are mutagenic BTW. Depending on amount and patient characteristics. Might be in only small amounts, but it is another argument against boosters and repeated dosing.
Here is the newest on N-1-methylpseudouridine and cancer. So are these nucleotides being recycled, plus extra unincorporated bases? Uh yeah, I think they knew about the risk of cancer but I will give the benefit of the doubt, maybe not in 2020.
Other general standard CQAs
I haven’t talke to much about bioburden, or endotoxin but I might mention this on the lipids CQA issues. But they now included a standard CQA for residual solvents. Oh like ethanol or the buffers HEPES? I will dig a little more.
Finally, what would the acceptable limits or amounts be? USP does not address this, only the regulators can. We are having discussions about the dsDNA, what about the aggregates, free bases, solvents, dsRNA etc etc.?
Summary
analytical methods are getting more sensitive and more specific and that is a good thing.
NEW impurities have been found in the mRNA THAT HAVE NOT BEEN FOUND BEFORE, because they didn’t know what was a quality impurity and what to test. Or not using sensitive enough assays.
THIS IS WHY YOU WANT A MATURE BIOLOGICAL PRODUCT FIRST, WITH ALL THE ANALYTICAL TESTING DONE before phase III trials.
and why, IMHO, Process 1, 2 and 3 are such a big deal.
this analysis is ONLY for the mRNA, when we get to the lipids, that’s when things get really interesting. Really interesting.
Everytime we look, we find more issues. Because these products are being transfected across membranes, they have to be 99.99999% pure. And at this point, I don’t think that is possible. (pharmacist tip: this is why oral therapy is almost always safer if you can get away with it, even with easy to purify IV drugs).
Pray the rosary







Two typos: accommodate, equipment.
There are several methods for many of these to accomadate differences in testing equipement
Excellent content...Dr Wiseman is a hero alongside of you. Thank you, as always.
Blame my editing/spelling on the nuns (also my penmanship, multiplication table)!
Brilliant summation, thank you for all the hard work, saving this as a reference!